NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data
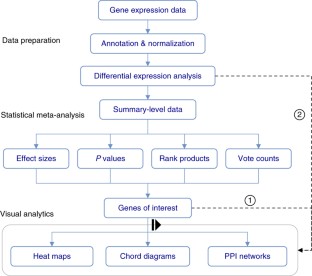
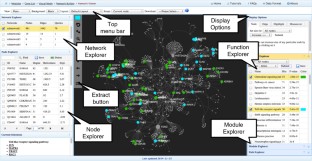
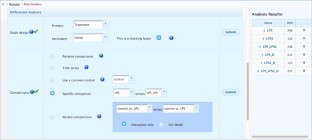
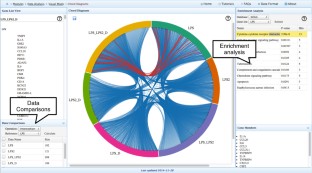
Meta-analysis of gene expression data sets is increasingly performed to help identify robust molecular signatures and to gain insights into underlying biological processes. The complicated nature of such analyses requires both advanced statistics and innovative visualization strategies to support efficient data comparison, interpretation and hypothesis generation. NetworkAnalyst (http://www.networkanalyst.ca) is a comprehensive web-based tool designed to allow bench researchers to perform various common and complex meta-analyses of gene expression data via an intuitive web interface. By coupling well-established statistical procedures with state-of-the-art data visualization techniques, NetworkAnalyst allows researchers to easily navigate large complex gene expression data sets to determine important features, patterns, functions and connections, thus leading to the generation of new biological hypotheses. This protocol provides a step-wise description of how to effectively use NetworkAnalyst to perform network analysis and visualization from gene lists; to perform meta-analysis on gene expression data while taking into account multiple metadata parameters; and, finally, to perform a meta-analysis of multiple gene expression data sets. NetworkAnalyst is designed to be accessible to biologists rather than to specialist bioinformaticians. The complete protocol can be executed in ∼ 1.5 h. Compared with other similar web-based tools, NetworkAnalyst offers a unique visual analytics experience that enables data analysis within the context of protein-protein interaction networks, heatmaps or chord diagrams. All of these analysis methods provide the user with supporting statistical and functional evidence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
265,23 € per year
only 22,10 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others

reString: an open-source Python software to perform automatic functional enrichment retrieval, results aggregation and data visualization
Article Open access 06 December 2021
NBIA: a network-based integrative analysis framework – applied to pathway analysis
Article Open access 06 March 2020

STAGEs: A web-based tool that integrates data visualization and pathway enrichment analysis for gene expression studies
Article Open access 02 May 2023
Accession codes
Accessions
Gene Expression Omnibus
Swiss-Prot
References
- Li, S. et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol.15, 195–204 (2014). ArticleGoogle Scholar
- Pena, O.M. et al. An endotoxin tolerance signature predicts sepsis and organ dysfunction at initial clinical presentation. EBioMedicine1, 64–71 (2014). ArticleGoogle Scholar
- Zhang, G. et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res.19, 4983–4993 (2013). ArticleCASGoogle Scholar
- Gieger, C. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet.4, e1000282 (2008). ArticleGoogle Scholar
- Gomez-Cabrero, D. et al. Data integration in the era of omics: current and future challenges. BMC Syst. Biol.8 (suppl. 2), I1 (2014). ArticleGoogle Scholar
- Tseng, G.C., Ghosh, D. & Feingold, E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res.40, 3785–3799 (2012). ArticleCASGoogle Scholar
- O'Donoghue, S.I. et al. Visualizing biological data-now and in the future. Nat. Methods7, S2–S4 (2010). ArticleCASGoogle Scholar
- Goble, C. & Stevens, R. State of the nation in data integration for bioinformatics. J. Biomed. Inform.41, 687–693 (2008). ArticleGoogle Scholar
- Smith, B. et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol.25, 1251–1255 (2007). ArticleCASGoogle Scholar
- Wodke, J.A. et al. MyMpn: a database for the systems biology model organism Mycoplasma pneumoniae. Nucleic Acids Res.43 (Database issue): D618–D623 (2014). PubMedPubMed CentralGoogle Scholar
- Breuer, K. et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res.41, D1228–D1233 (2013). ArticleCASGoogle Scholar
- Rhodes, D.R. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia9, 166–180 (2007). ArticleCASGoogle Scholar
- Chelaru, F., Smith, L., Goldstein, N. & Bravo, H.C Epiviz: interactive visual analytics for functional genomics data. Nat. Methods11, 938–940 (2014). ArticleCASGoogle Scholar
- Nekrutenko, A. & Taylor, J. Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat. Rev. Genet.13, 667–672 (2012). ArticleCASGoogle Scholar
- Xia, J. et al. INMEX—a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res.41, W63–W70 (2013). ArticleGoogle Scholar
- Xia, J., Lyle, N.H., Mayer, M.L., Pena, O.M. & Hancock, R.E.W. INVEX—a web-based tool for integrative visualization of expression data. Bioinformatics29, 3232–3234 (2013). ArticleCASGoogle Scholar
- Xia, J., Benner, M.J. & Hancock, R.E.W. NetworkAnalyst—integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res.42, W167–W174 (2014). ArticleCASGoogle Scholar
- Tarraga, J. et al. GEPAS, a web-based tool for microarray data analysis and interpretation. Nucleic Acids Res.36, W308–W314 (2008). ArticleCASGoogle Scholar
- Reich, M. et al. GenePattern 2.0. Nat. Genet.38, 500–501 (2006). ArticleCASGoogle Scholar
- Xia, J., Mandal, R., Sinelnikov, I.V., Broadhurst, D. & Wishart, D.S. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res.40, W127–W133 (2012). ArticleCASGoogle Scholar
- Xia, J. & Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc.6, 743–760 (2011). ArticleCASGoogle Scholar
- Saeed, A.I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques34, 374–378 (2003). ArticleCASGoogle Scholar
- Perez-Llamas, C. & Lopez-Bigas, N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE6, e19541 (2011). ArticleCASGoogle Scholar
- Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). ArticleCASGoogle Scholar
- Huang, D.W. et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res.35, W169–W175 (2007). ArticleGoogle Scholar
- Reimand, J., Arak, T. & Vilo, J. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res.39, W307–W315 (2011). ArticleCASGoogle Scholar
- Lynn, D.J. et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol.4, 218 (2008). ArticleGoogle Scholar
- Saito, R. et al. A travel guide to Cytoscape plugins. Nat. Methods9, 1069–1076 (2012). ArticleCASGoogle Scholar
- Smyth, G.K. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor (eds. Gentleman, R. et al. 397–420 (Springer, 2005).
- Law, C.W., Chen, Y., Shi, W. & Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol.15, R29 (2014). ArticleGoogle Scholar
- Orchard, S. et al. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat. Methods9, 345–350 (2012). ArticleCASGoogle Scholar
- Turinsky, A.L., Razick, S., Turner, B., Donaldson, I.M. & Wodak, S.J. Interaction databases on the same page. Nat. Biotechnol.29, 391–393 (2011). ArticleCASGoogle Scholar
- Bader, G.D., Betel, D. & Hogue, C.W. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res.31, 248–250 (2003). ArticleCASGoogle Scholar
- Stark, C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res.34, D535–D539 (2006). ArticleCASGoogle Scholar
- Licata, L. et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res.40, D857–D861 (2012). ArticleCASGoogle Scholar
- Hermjakob, H. et al. IntAct: an open source molecular interaction database. Nucleic Acids Res.32, D452–D455 (2004). ArticleCASGoogle Scholar
- Rolland, T. et al. A proteome-scale map of the human interactome network. Cell159, 1212–1226 (2014). ArticleCASGoogle Scholar
- Pena, O.M., Pistolic, J., Raj, D., Fjell, C.D. & Hancock, R.E.W. Endotoxin tolerance represents a distinctive state of alternative polarization (M2) in human mononuclear cells. J. Immunol.186, 7243–7254 (2011). ArticleCASGoogle Scholar
- Ramasamy, A., Mondry, A., Holmes, C.C. & Altman, D.G. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med.5, e184 (2008). ArticleGoogle Scholar
- Mitra, K., Carvunis, A.R., Ramesh, S.K. & Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet.14, 719–732 (2013). ArticleCASGoogle Scholar
- Ideker, T., Ozier, O., Schwikowski, B. & Siegel, A.F. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics18 (suppl. 1), S233–S240 (2002). ArticleGoogle Scholar
- Beisser, D., Klau, G.W., Dandekar, T., Muller, T. & Dittrich, M.T. BioNet: an R package for the functional analysis of biological networks. Bioinformatics26, 1129–1130 (2010). ArticleCASGoogle Scholar
- Vidal, M., Cusick, M.E. & Barabasi, A.L. Interactome networks and human disease. Cell144, 986–998 (2011). ArticleCASGoogle Scholar
- Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Science322, 104–110 (2008). ArticleCASGoogle Scholar
- Schramm, S.J. et al. Disturbed protein-protein interaction networks in metastatic melanoma are associated with worse prognosis and increased functional mutation burden. Pigment Cell Melanoma Res.26, 708–722 (2013). ArticleCASGoogle Scholar
- Liu, Y., Koyuturk, M., Barnholtz-Sloan, J.S. & Chance, M.R. Gene interaction enrichment and network analysis to identify dysregulated pathways and their interactions in complex diseases. BMC Syst. Biol.6, 65 (2012). ArticleCASGoogle Scholar
- Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res.19, 1639–1645 (2009). ArticleCASGoogle Scholar
- Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. in International AAAI Conference on Weblogs and Social Mediahttp://www.aaai.org/ocs/index.php/ICWSM/09/paper/viewFile/154Forum/1009 (2009).
Acknowledgements
The authors thank the Canadian Institutes for Health Research (CIHR) for financial support.
Author information
Authors and Affiliations
- Department of Microbiology and Immunology, University of British Columbia, Vancouver, British Columbia, Canada Jianguo Xia, Erin E Gill & Robert E W Hancock
- and Department of Animal Science, Institute of Parasitology, McGill University, Ste. Ann de Bellevue, Québec, Canada Jianguo Xia
- Department of Microbiology and Immunology, McGill University, Montreal, Québec, Canada Jianguo Xia
- Wellcome Trust Sanger Institute, Hinxton, United Kingdom Robert E W Hancock
- Jianguo Xia












